@cmancuso and I finally got around to doing the calibration with two photodiodes. I rewrote the code on the server, electron GUI, and the calibration script to simplify some of the data processing and allow us to simultaneously calibrate both photodiodes at once.
Device Settings
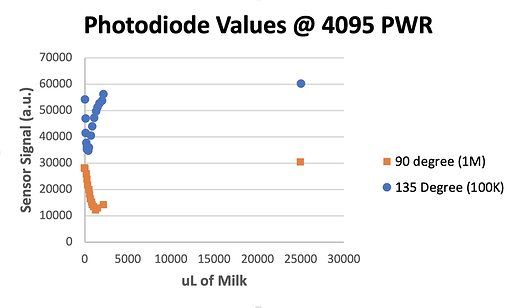
Resistor Pack OD90: 1M
Resistor Pack OD135: 100K
OD90 Averaging: 1000
OD135 Averaging: 1000
LED Power: 4095 (full power)
Calibration Graphs
OD90
OD135
I also wrote code to generate 3D subplots, but they weren’t very informative to look at - I think looking at the two individual sets of calibration curves provides more insight to the data. I put some information at the bottom in anyone is interested.
Thoughts
@cmancuso and I discussed this for a while, and please anyone feel free to add any comments or thoughts you might have from looking at these graphs.
The first thing that was nice to see is that the E. Coli curves match the curves @bgwong provided using milk. For 135 degrees, there is a steep increase in signal (decrease in ADC value) initially as the cells or milk scatter the light into the PD, followed by a steady decline of signal as they become more dense and block out the light completely. The 90 degree curves show a similar trend, but are more stretched out as the angle is much more intense.
We noticed that curves that were particularly bad for the 90 degree at the low OD range (like vials 0, 2, and 14) were excellent at low OD for the 135 degree PD. Vials that the 90 degree PD is able to measure at low ODs (like vial 8) seem to have 135 degree values that are already being pushed into the regime where the signal is decreasing as density increases, meaning the scattering signal is saturated for that angle.
@bgwong can correct me if I get some of this incorrect, but from my understanding, increasing the value of the resistor pack means that less current (or a smaller signal) is able to generate a larger voltage drop on the ADC (up to 3.3 V - 0.6 V dropped across the diode). This means that you can get really good signal from less photons hitting the detector. This is why the NBT paper used a larger resistor pack (10M) with a lower LED power (2125). The problem is that you can saturate the signal rather quickly as the voltage will not increase over the bias voltage on the PD minus the drop over the PD. So in the new setups we’ve been trying, we use smaller resistor packs with more LED to try and get a larger dynamic range.I think now we are seeing not the sensor saturating, but the light scattering from the cells saturating, which is kind of cool. By having the dual angle/resistor pack, it’s possible to get really nice data from lower OD cultures via the 135 degree photodiode, while still being able to measure higher ODs with the 90 degree and shift in the 135 from sensing scattering to being blocked out.
Going forward
There are a few options here that anyone can do with their units. The first is to go the NBT route - tune the LED power and resistor pack to give a range that works for whatever system being studied. A resistor pack of 10M and LED power of 2125 at 135 degrees gives pretty good results for most vials from OD 0.0 0.7, give or take a few tenths depending variability in the LED, PD, and their relative positions in the 3d printed part. You could also use the 90 degree PD if you’d like to explore higher ODs, but you might lose some of the lower end accuracy, which might not matter too much depending on the application.
@cmancuso and I also discussed possibly using both PDs in conjunction to determine the OD of the culture. The simplest way to do this would be to select the curve that has the steepest slope at the value being measured. This could introduce noisy measurements at points where the slopes are nearly identical or switching between the curves if they do not exactly align. There could be a way to weight the predicted OD from each curve based on their relative slopes, but we’d need to test this out to see how it would work in practice.
Also, as @bgwong mentioned we could use a 3d surface fit, but again this would need to be tested out to really see if it will work. I uploaded my calibration data here. You need to use the new version of the calibration script here. I’ll update the link when it’s merged to the main branch. The script takes an argument --3d if you want to plot in 3d using both 135 and 90 degree measurements.
I’m really not sure about the feasibility of using a 3d surface fit for calibrations, I think there might be a clever way to maybe using some machine learning methods to use both the 90 and 135 data to determine OD. Maybe @mgalardini could chime in here.